Interesting sex chromosomes in Mercurialis annua
The Y chromosome of Mercurialis annua is homomorphic yet 1/3 is not recombining and is enriched in sex-biased genes, perhaps a sign of ongoing sexual antagonism.

Most of the times when you see a plant with separate male and female individuals, you are witnessing an example of a new evolution of sex chromosomes.
The study of different plant species is therefore a great opportunity to test the theoretical predictions about how sex chromosomes evolve and become different to each other. There are non-selective and selective reasons for this. The former involve their smaller population size compared to the autosomes, while the latter come down to the fact that each sex chromosome experiences a different genomic and ecological environment, which is specific to each sex. To be exact, there is 100% association of the Y chromosome and 33% association of the X chromosome with males (and 66% association with females, since they have two copies of the X).
Old sex chromosomes (of mammals, birds, Drosophila and some plants) have been found to be genetically degenerated. This is predicted to be a secondary consequence of the evolution of recombination suppression on the sex chromosomes, which can only be studied in young sex chromosomes, because the degeneration of old sex chromosomes masks most of the beneficial effects associated with each, that are hypothesised to have been the reason for the suppression of recombination.
In principle, the sex chromosomes are expected to stop recombining if this leads to the generation of combinations of genes that benefit the sex in which the sex chromosomes occur most frequently (the Y in male and the X in females). For example, a male-beneficial variant linked to the male-determining gene will achieve a higher frequency in the population relative to an alternative Y chromosome that sometimes recombines to lose this variant. Similarly, a Y chromosome carrying a male-beneficial gene variant that is simultaneously deleterious to females will outcompete other Y variants even faster, because the products of recombination will be outcompeted even faster. These genetic variants that have opposite fitness consequences for the different sexes are termed "sexually antagonistic".
In the short term the lack of recombination between sex chromosomes carrying sexually antagonistic genes maximises the fitness of each sex, so they are expected to accumulate on the sex chromosomes. However, the recombination of genetic information is beneficial over the long term, for example it allows the removal of deleterious mutations that occur within a stretch of genes. That is why the fate of all non-recombining sex chromosomes is to degenerate.
And this brings us to the recently published genome of the annual plant M. annua. We seem to have been very lucky in choosing M. annua as a study species because it shows signs of recombination suppression without much of the degeneration that is expected to follow. Its sex chromosomes may therefore be in the right stage where the reason for recombination suppression (sexually antagonistic selection) may be still present, and not masked by degeneration.
The M. annua genome project evolved over a long period of time and combined lots of different datasets. We report an assembly of the genome, a genetic map that assigned ≈10,000 genes on the expected number of chromosomes and a transcriptome that allowed to identify genes that are expressed at a higher level in one sex relative to the other (sex-biased genes). We also obtained various metrics associated with DNA sequence, such as divergence to other species or within-species diversity and tested whether the sex chromosomes stand out in any of these metrics. Having a genome allowed to produce a high throughput assay allowing to selectively sequence genes in many populations covering the species range.
Our genetic map and species-wide data independently confirm that 1/3 of the sex chromosomes do not recombine. This is surprising because the recombination suppression was recent: we found only one clear case of a non-functional Y copy and comparisons with the closely related species M. huetii showed that the recombination suppression happened after the species split.
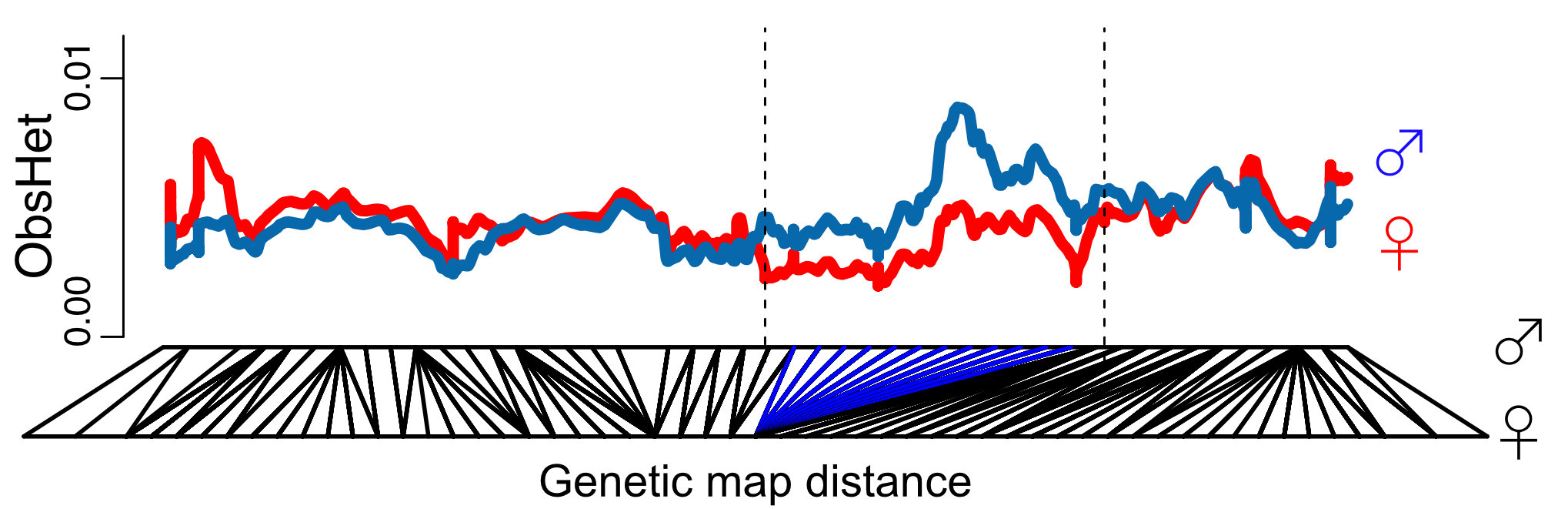
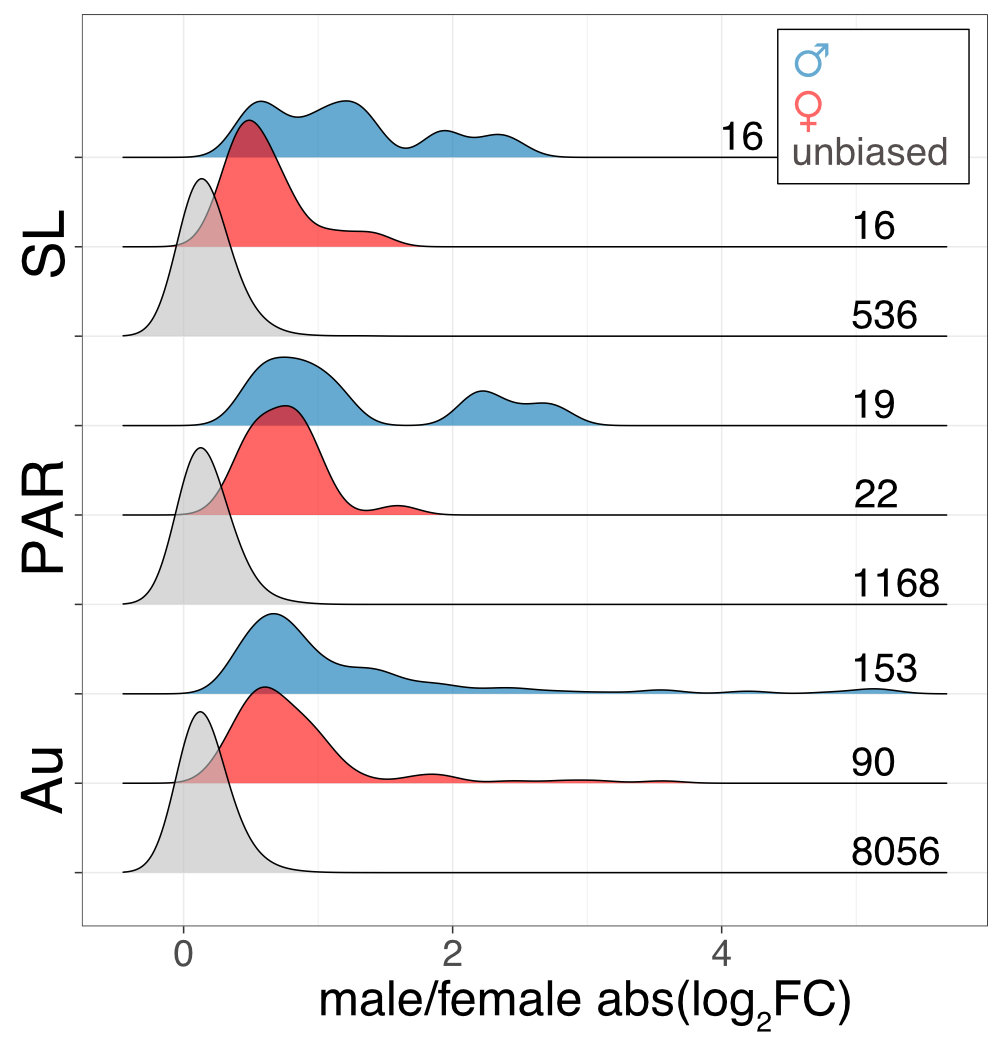
Interestingly the sex chromosomes are enriched in sex-biased genes. Overall there are more genes with higher expression in females than males, but the few genes with higher expression in males compared to females have more extensive expression differences between the sexes. Assuming that sex bias in gene expression is associated with fitness, which would have caused the lower expression in the opposite sex, this simultaneous masculinisation and feminisation of the sex chromosome in terms of gene expression can be interpreted as the outcome of ongoing sexual conflict. It may have been associated with the reason behind the observed recombination suppression between the sex chromosomes.
This pattern is only a hint towards potential ongoing sexually antagonistic selection on the sex chromosomes, which needs to be followed up. The existence of a large region with suppressed recombination without much degeneration, and the availability of many closely related lineages for comparative genomic analysis makes M. annua a very promising species to understand the early events in sex chromosome evolution.
Read the full paper here.